Rapid preparation method of plant genomic DNA for PCR Source: Fangtong Technology
-------------------------------------------------- --------------------------------
Abstract: This paper describes a rapid preparation method for plant genomic DNA for PCR. In this method, a small amount (4-6 mg) of plant leaves, an appropriate amount (200 μl) of TE buffer solution, and a tungsten alloy bead are placed in a centrifuge tube, and after shaking the tissue for 5 min in a tissue cell grinder, 1 μl of the DNA solution is directly taken. As a template for PCR. The method has the advantages of simple and rapid, low cost, good amplification effect, and the like, and is particularly suitable for genotype detection of a large number of samples.
Wang Lan 1, 2 Long Yun Ming 2 Liu Yaoguang 2
1 College of Agriculture, South China Agricultural University, Guangdong Key Laboratory of Plant Molecular Breeding
2 College of Life Sciences, South China Agricultural University, Key Laboratory of Plant Functional Genomics and Biotechnology, Guangdong Provincial Department of Education
Abstract: This paper describes a rapid method for the preparation of plant genomic DNA for PCR. In this method, a small amount (4-6 mg) of plant leaves, an appropriate amount (200 μl) of TE buffer solution, and a tungsten alloy bead are placed in a centrifuge tube, and after shaking the tissue for 5 min in a tissue cell grinder, 1 μl of the DNA solution is directly taken. As a template for PCR. The method has the advantages of simple and rapid, low cost, good amplification effect, and the like, and is particularly suitable for genotype detection of a large number of samples.
Keywords: rice, DNA preparation, PCR
With the rapid development of biotechnology, PCR technology has been widely used in various fields of genetic breeding, such as seed purity identification, phylogenetic analysis, molecular marker-assisted breeding, gene mapping, and transgenic plant detection. Efficient and rapid template DNA preparation is very important in large-scale PCR-based genotyping. Since Marmur established the classical method for the isolation and extraction of DNA samples from biological cells with sodium dodecyl sulfate (SDS) and chloroform in 1961, people have been working on DNA extraction methods and have reported some DNA. Extraction method (Dellaporta et al., 1983; Xuan Pu et al., 1998; Wang Xiufeng et al., 2002; Zhou Shufen et al., 2004). But these methods are still cumbersome and time consuming. In this study, the preparation method of plant genomic DNA as a PCR template was simplified. Only a small amount of leaves, an appropriate amount of TE buffer, and a tungsten alloy bead were placed in a centrifuge tube, and the tissue was shaken in a tissue cell grinder for about 5 minutes to break the tissue. Directly take 1 μl of DNA solution as a template for PCR. The method has the advantages of simple and rapid, low cost, good amplification effect, and the like, and is particularly suitable for large-scale genotype detection.
1 Materials and methods
1.1 Test materials The test materials were the indica line E5 of indica variety Taizhong 65 (T65) and its hybrid sterility gene Sc, and the F2 population constructed by their hybrid combination, and the Arabidopsis thaliana ecotype Columbia.
1.2 Preparation method of genomic DNA (take or remove by hand) rice seedlings or adult leaves (cut into about 3 ~ 5mm small section) or Arabidopsis leaves about 4 ~ 6mg, 12 ~ 15mg, or 25 ~ 30mg, put In a 2ml centrifuge tube (Note: the bottom of the 2ml centrifuge tube is wider, which is good for the oscillation of the tungsten alloy beads), add 200μl TE (10mmol/L Tris, pH 8.0; 0.5mmol/L EDTA, pH 8.0. Note: This TE The concentration of EDTA is 1/2 of conventional TE or 200 μl of deionized water, and a 3 mm diameter tungsten alloy bead (Qiagen, Cat. 69997) is placed in a multi-functional tissue cell grinder (Fangtong Biotechnology Co., Ltd.) , model MTM-60) vibrates for about 5 minutes to break the blade. 17 μl of the solution was taken to detect the mass of the template DNA by agarose gel electrophoresis, and 1 μl of the solution was taken for PCR amplification.
1.3 PCR Primer Reaction PCR primers were designed according to the rice and Arabidopsis genome sequences of the public database (Table 1), synthesized by Handsome. Taq enzyme was purchased from Jieshun Company.
Table 1 PCR primer sequences
| Primer | Primer sequence |
| PR1 | 5'-TCTGTACAAGTTGAGGAATCTCCG-3' |
| Â | 5'-TGCCACCCACAGCAAGCCTTATGA-3' |
| PR2 | 5'-ACCCCATGGCGACGAATTCC-3' |
| Â | 5'-CAGATTAAGGCAGGAAGTCC-3' |
| PA | 5'-GAAACTGATCAGTTGCTTTCAC-3' |
| Â | 5'-CTGCTACACAATCTCGTTATGGAG-3' |
| L14-121 | 5'-CTTGACAATTGCAAGGCCAA-3' |
| Â | 5'-GAGGTAGGCATGAGACATGC-3' |
| J04-91 | 5'-GTGCGGATCGATCGGCAGCA-3' |
| Â | 5'-GCTCAGATCCGGCCAGATTC-3' |
| P24-83 | 5'-CAGAACAGCATTCAGGCATC-3' |
| Â | 5'-ATCCTGATAATGTGTGGCGC-3' |
| P24-93 | 5'-CATTGGTTAAAGGATGGTAC-3' |
| Â | 5'-TAGTACTGCTAGGACCATCC-3' |
Each PCR reaction solution (20 μl) was composed of: 1 × Buffer, 0.2 mmole/L dNTPs, 1 U Taq enzyme, 250 nmole/L of each primer, and 1 μl of the above DNA solution. The PCR reaction was carried out on a My Cycler (BioRad) thermal cycler. Pre-denaturation was carried out at 94 ° C for 3 min using a primer reaction procedure, and amplification was carried out for 35 cycles of 94 ° C for 20 s, 55 to 58 ° C for 30 s, 72 ° C for 60 s (InDel-labeled PCR for 30 s), and finally 72 ° C for 2 min.
1.4 Polyacrylamide gel electrophoresis and detection
1.4.1 Preparation of polyacrylamide gel Prepare 6% polyacrylamide gel 100ml working solution: add 5.7 g acrylamide (Acri), 0.3 g N, N-methylene bis acrylamide (Bis-Acri) , 12 g of urea, 10 ml of 10 x TBE, and finally add water to 100 ml. Before filling the gel, add 80μl of 10% ammonium persulfate and 4μl of tetramethylethylenediamine (TEMED) per 10ml of glue. After shaking, quickly fill the gel. After the gel is solidified, it can be spotted and electrophoresed. .
1.4.2 Silver staining is first stained with 0.1% AgNO3 staining solution for 10~15min, and then developed with developer solution (0.5mL containing NaOH, 100g sodium tetraborate, 0.019g, 0.4mL formaldehyde). After the strip is clear, use it directly. Tap water rinse to stop the color reaction, photograph with a gel imager (BioRad) or a digital camera and record the results.
2 Results and discussion
2.1 Rapid preparation of PCR template DNA This method is to directly obtain a DNA solution by shaking a small amount of plant leaves in TE solution or deionized water through the shaking of tungsten alloy beads in a centrifuge tube. Sixty samples were operated at a time using a multifunctional tissue cell grinder (MTM-60). Agarose gel electrophoresis showed that a larger molecular weight DNA fragment was obtained with both TE and deionized water (Fig. 1A). The DNA prepared by TE can be stored at 4 ° C for more than 10 days and can be stored for half a year under freezing. The DNA prepared with deionized water is easily degraded and has a short storage time. Therefore, we mainly use TE.

Figure 1. Rice and Arabidopsis genomic DNA prepared by this method.
Note: A: Rice genomic DNA (4-6 mg leaves/200 μl) prepared using TE (lanes 1 to 5) and deionized water (lanes 6 to 10). M is a molecular weight marker. B, C: Different concentrations of rice (B) and Arabidopsis (C) genomic DNA prepared with 200 μl of TE. Lanes 1-3 are 4-6 mg leaves; 4-6 are 12-15 mg leaves; 7-9 are about 25-30 mg leaves.
2.2 PCR amplification of different amounts of template DNA The genomic DNA prepared by this method was not purified and contained various impurities which may inhibit the activity of Taq enzyme. Therefore, adding too much crude DNA may affect the PCR reaction. To find the amount of DNA suitable for the PCR reaction, we added different amounts (4-6 mg, 12-15 mg, and 25-30 mg) of rice and Arabidopsis leaves to prepare genomic DNA in a certain amount of TE (200 μl) (Figure 1 , B, C). Two pairs of rice primers (PR1, PR2, amplification products of 540 bp and 890 bp, respectively) and one pair of Arabidopsis primers (PA, amplification product length of 1 kb) were selected, and 1 μl of DNA was subjected to PCR reaction in 20 μl system. . The results showed that for the amplification of smaller DNA fragments, all templates amplified the product, but the template DNA prepared with 4-6 mg leaves obtained the best amplification effect (Fig. 2A). For amplification of larger DNA fragments, only template DNA prepared with 4-6 mg leaves obtained a more stable amplification effect (Fig. 2B, C).
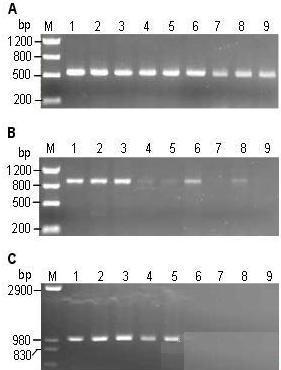
Figure 2 Agarose gel (1%) detection of different amounts of genomic DNA as a template for PCR reaction
Note: A, B, and C are PCRs using primers PR1, PR2, and PA, respectively. In all reactions (20 μl) containing 1 U Taq enzyme and 1 μl genomic DNA; Lanes 1-3, 4-6, 7-9 were genomes prepared in 200 μl TE with 4-6 mg, 12-15 mg, and 25-30 mg leaves, respectively. DNA.
This experiment demonstrates that the preparation of the genomic DNA solution with a ratio of 4 to 6 mg (up to about 10 mg) of the leaves/200 μl of TE has a low impurity concentration, and the addition of 1 μl of the DNA solution to the 20 μl reaction system does not inhibit the activity of the Taq enzyme. The addition of 1 μl of higher concentration of template DNA has a greater inhibitory effect on the PCR reaction. Since the amount of template DNA is small (about 1 to 2 ng/μl), the number of PCR cycles is more than that of a larger amount of purified DNA template (usually 20 to 50 ng) (more than 3 to 5 cycles). Although it is possible to dilute template DNA prepared with more leaves or add a smaller amount (<1 μl) of template DNA to obtain good PCR results, but to simplify the operation steps and improve work efficiency, determine the best empirical value. It is especially important for efficient large-scale sample testing. Since there is a suitable range of the amount of sample leaves to be added, in practice, it is not necessary to use all the scales to be weighed by a balance, and it can be estimated by experience.
2.3 Genotypic analysis for molecular markers Since the genomic DNA prepared by this method can be PCR-amplified for larger target fragments, for PCR molecular markers such as microsatellite (SSR), insertion/deletion (InDel), single nucleoside Detection of shorter DNA fragments (generally <200 bp) such as acid polymorphism (SNP) should be feasible. We used four pairs of InDel-labeled primers (Table 1) located in the linkage region of the rice 籼粳 hybrid pollen sterility gene Sc (Yang et al., 2008) to perform genotypic analysis of the F2 population. Figure 3 shows partial results. The PCR amplification of all markers is stable, and the DNA bands of polyacrylamide gel electrophoresis are clearly distinguishable.

Figure 3 Detection of rice genotypes labeled with InDel
Note: A to D are PCRs using primers L14-121, J04-91, P24-83, and P24-93, respectively. Lanes 1 and 2 on the left are parental E5 and T65, respectively, and all other lanes are F2 individuals.
A number of methods have been reported for the preparation of plant genomic DNA, such as the classical SDS method, the CTAB method, and some simplified preparation methods, such as the SDS one-step method (Wang Huiwen et al., 2002), the microwave boiling method (Huang Xiaole et al., 2002), TE grinding method (Luo Wenyong et al., 2002), NaOH treatment of young leaves directly as a PCR template method (Wang Xiufeng et al., 2002), simple extraction method (Chen Wenyue et al., 2005), and shearing method (Zhao Hongxia et al., 2006). For studies that require genotyping of a large number of samples, the efficiency of these methods is not high enough, or the amplification of the prepared template DNA is not ideal, and the target fragment is often not amplified. The DNA preparation method proposed in this study is simple, greatly shortens the time, is suitable for operating a large number of samples, and has good PCR effect and repeatability, requires a small amount of leaves, and the required instrument (multi-functional tissue cell grinder) is low in price. Therefore, the cost of the experiment is low. We have used this method to prepare tens of thousands of rice samples for genotyping of various molecular markers (SSR, InDel, SNP) and transgenes.
Integrated Mask Manufacturing Machine
Guangzhou Aikangli Medical Technology Co., Ltd. , https://www.aikanli.com